
INSILICO VACCINE SUITE
InSilicoVACCINE
Computational tools can play a vital role in streamlining and de-risking vaccine development to achieve a faster process, lower costs, and higher vaccine efficacy.

INSILICO VACCINE SUITE
Computational tools can play a vital role in streamlining and de-risking vaccine development to achieve a faster process, lower costs, and higher vaccine efficacy.

Vaccine development is a highly challenging process which requires technically complex and expensive vaccine design and aims to universally weaponize a heterogenous populations’ variable immune systems against continually evolving pathogens. Computational tools can play a vital role in streamlining and de-risking vaccine development to achieve a faster process, lower costs, and higher vaccine efficacy.
Our InSilicoVaccine suite combines immunoinformatic tools and immune system response predictions that can support early vaccine design with disease progression simulations and virtual populations that allow evaluation of clinical efficacy. The heterogeneous virtual populations can be applied to test treatment strategies in silico – both if the vaccine is administered solo and when it is part of a combination therapy – and define target populations. The InSilicoVaccine suite pipeline provides a broad range of tools that enable analysis of the vaccine’s efficacy and support development throughout the vaccine’s life cycle.
Influenza A
Our InSilicoVaccine pipeline can support vaccine design for Influenza A throughout the development pipeline, through its innovative combination of immunoinformatics tools with disease modeling and virtual populations. In particular, the integration of not only the systemic immune response, but also a specific disease model of influenza A, allows predictions of vaccine efficacy in an infection.
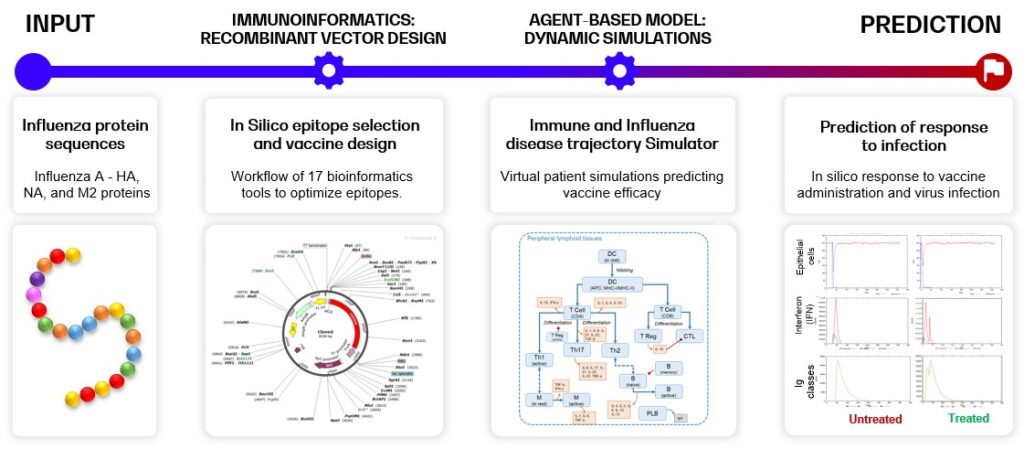
Below, an example of multi-epitope recombinant vector vaccine design supported by the InSilicoVACCINE pipeline with the Influenza A disease model is shown. The pipeline combines a variety of tools and capabilities, and consists of two major steps:
Influenza A
COVID-19
Computational tools show great potential to play a key role in the fast and cost-effective design and evaluation of vaccines and treatments [2]–[4]. Our InSilicoVACCINE suite can be used not only for de novo vaccine design, but also to identify and evaluate the effectivity of existing vaccine components in as additives to novel vaccines.
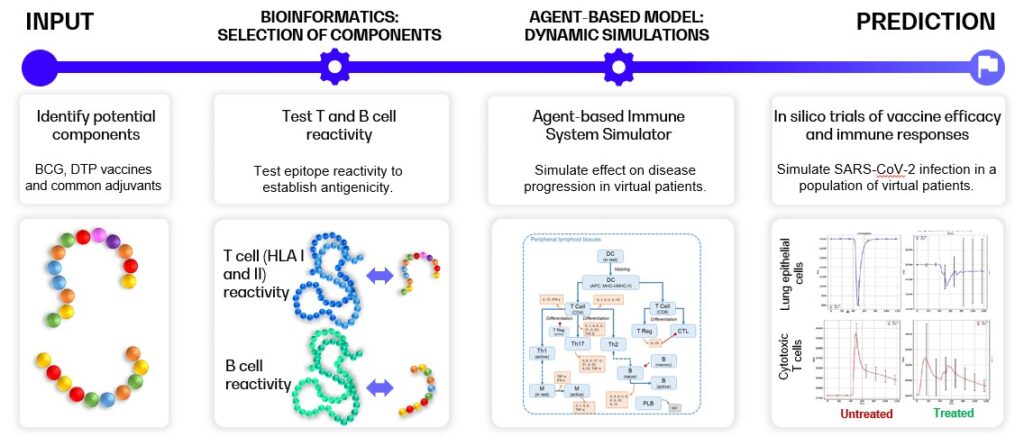
Below, an example of selection and evaluation of components for future COVID-19 vaccines is shown [5], [6]. The workflow consists of several distinct steps: (1) identification of elements of BCG and DTP vaccines and selected vaccines adjuvants that show similarity with the SARS-CoV-2 genome, (2) testing of the antigenicity of these identified epitopes by in silico prediction of the T and B cell receptor reactivities, and (3) in silico trials of the vaccines components’ effects on COVID-19 infection through heterogeneous virtual patients of COVID-19 disease course.
Influenza A
Tuberculosis
Our InSilicoVACCINE pipeline can streamline the clinical development of novel tuberculosis treatments by supporting trial design, dosing and scheduling of combination therapies, and regulatory pathways with in silico trials of tuberculosis treatment.
Key components of InSilicoVACCINE are disease progression models and virtual populations. These modelling techniques allow not only a detailed analysis of how treatment may affect active or latent infections, but also allows deeper insight into which immune system components are, or are not, incorporated in the protective mechanism, and how each immune system mechanism can be optimally leveraged.
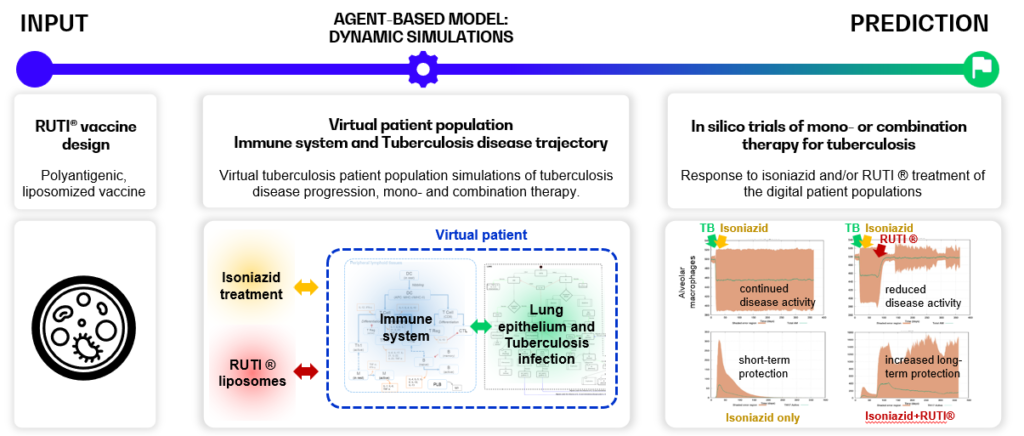
Below, an example of in silico trials for the RUTI ® vaccine – a polyantigenic vaccine, consisting of liposomized Mycobacterium Tuberculosis fragments, is shown. The in silico trials show (combination) therapy efficacy, and can support dosing and administration schedule design to optimize vaccine efficacy [4]–[7].
Influenza A
[formidable id=”14″]